Photometry with XGA
This tutorial will show you how to perform basic photometric analyses on XGA sources, starting with how you use the built in SAS interface to generate the necessary XMM images, exposure maps, and the XGA ratemap objects. Using the SAS interface requires that a version of SAS be installed on your system, and the module will check for the presence of this software before trying to run any SAS routines.
I will take you through the basics of interacting with the XGA photometric products, including using source masks and the built in peak finding techniques.
[1]:
from astropy.units import Quantity
from astropy.visualization import LinearStretch
import numpy as np
import pandas as pd
from xga.sources import GalaxyCluster, NullSource
from xga.samples import ClusterSample
from xga.sas import evselect_image, eexpmap, emosaic
from xga.utils import xmm_sky
First of all, I will declare an individual galaxy cluster source object, and a galaxy cluster sample object. I am going to demonstrate that you can use the SAS interface with individual sources and samples of sources in exactly the same way. The sample of four clusters is taken from the XCS-SDSS sample (Giles et al. (in prep)), and the individual cluster is Abell 907.
[2]:
# Setting up the column names and numpy array that go into the Pandas dataframe
column_names = ['name', 'ra', 'dec', 'z', 'r500', 'r200', 'richness', 'richness_err']
cluster_data = np.array([['XCSSDSS-124', 0.80057775, -6.0918182, 0.251, 1220.11, 1777.06, 109.55, 4.49],
['XCSSDSS-2789', 0.95553986, 2.068019, 0.11, 1039.14, 1519.79, 38.90, 2.83],
['XCSSDSS-290', 2.7226392, 29.161021, 0.338, 935.58, 1359.37, 105.10, 5.99],
['XCSSDSS-134', 4.9083898, 3.6098177, 0.273, 1157.04, 1684.15, 108.60, 4.79]])
# Possibly I'm overcomplicating this by making it into a dataframe, but it is an excellent data structure,
# and one that is very commonly used in my own analyses.
sample_df = pd.DataFrame(data=cluster_data, columns=column_names)
sample_df[['ra', 'dec', 'z', 'r500', 'r200', 'richness', 'richness_err']] = \
sample_df[['ra', 'dec', 'z', 'r500', 'r200', 'richness', 'richness_err']].astype(float)
# Defining the sample of four XCS-SDSS galaxy clusters
demo_smp = ClusterSample(sample_df["ra"].values, sample_df["dec"].values, sample_df["z"].values,
sample_df["name"].values, r200=Quantity(sample_df["r200"].values, "kpc"),
r500=Quantity(sample_df["r500"].values, 'kpc'), richness=sample_df['richness'].values,
richness_err=sample_df['richness_err'].values)
# And defining an individual source object for Abell 907
demo_src = GalaxyCluster(149.59209, -11.05972, 0.16, r500=Quantity(1200, 'kpc'), r200=Quantity(1700, 'kpc'),
name="A907")
Declaring BaseSource Sample: 100%|██████████| 4/4 [00:01<00:00, 2.77it/s]
Generating products of type(s) ccf: 100%|██████████| 4/4 [00:10<00:00, 2.70s/it]
Generating products of type(s) image: 100%|██████████| 4/4 [00:03<00:00, 1.32it/s]
Generating products of type(s) expmap: 100%|██████████| 4/4 [00:01<00:00, 2.24it/s]
Setting up Galaxy Clusters: 100%|██████████| 4/4 [00:03<00:00, 1.23it/s]
A note on XGA’s Parallelism
In every SAS function built into XGA (and indeed many functions in other parts of the module), you will find a num_cores keyword argument which tells the function how many cores on your local machine it is allowed to use. The default value is 90% of the available cores on your system, though you are of course free to set your own value when you call the functions.
To see the number of cores which have automatically allocated to XGA, you can import the NUM_CORES constant from the base xga module (this tutorial for instance was run on a laptop with an Intel Core i5-8300H CPU, with four physical cores and four logical cores):
[3]:
from xga import NUM_CORES
NUM_CORES
[3]:
7
You can also manually set this value globally, before running anything. Either set the num_cores option in the [XGA_SETUP] section of the configuration file, or simply set the NUM_CORES constant imported from xga.
Generating XMM Images with XGA
Please note that the XCS data reduction pipeline produces images and exposure maps in the 0.5-2.0keV and 2.0-10.0keV energy ranges, and as the configuration file points XGA towards existing data, these sources will already have those images and exposure maps loaded in.
Please also note that ClusterSample class automatically generates images and exposure maps in the energy range used for peak finding (set by the keyword arguments peak_lo_en and peak_hi_en, by default 0.5keV and 2.0keV respectively). An individual GalaxyCluster source object will also automatically generate images and exposure maps in this range, but only if the use_peak keyword argument is set to True (which it is by default).
As such, I’m going to demonstrate how you can generate images and exposure maps in another (probably not very useful in a real analysis) energy range, 0.5-2.0keV. When telling SAS what energy range to generate in, we use Astropy quantities in units of keV, though you could supply limits in any energy unit:
[4]:
# Setting up the lower and upper energy bounds for our products
lo_en = Quantity(0.5, 'keV')
hi_en = Quantity(10.0, 'keV')
[5]:
# This will generate images in the 0.5-10keV range, for every instrument in
# every ObsID in every source in this sample
demo_smp = evselect_image(demo_smp, lo_en=lo_en, hi_en=hi_en)
# The function call is identical when generating images for an individual source, but here
# I have limited XGA to using only four cores
demo_src = evselect_image(demo_src, lo_en=lo_en, hi_en=hi_en, num_cores=4)
Generating products of type(s) image: 100%|██████████| 12/12 [00:15<00:00, 1.31s/it]
Generating products of type(s) image: 100%|██████████| 9/9 [00:04<00:00, 1.97it/s]
These images have now been generated and associated with the appropriate source.
Generating XMM Exposure Maps with XGA
The process for generating exposure maps is almost identical to the image generation process, we simply call a different function, though XMM calibration files (CCFs) are required to make exposure maps, so they will be generated if they do not already exist:
[6]:
# This will generate expmaps in the 0.5-10keV range, for every instrument in
# every ObsID in every source in this sample
demo_smp = eexpmap(demo_smp, lo_en=lo_en, hi_en=hi_en)
# The function call is identical when generating expmaps for an individual source
demo_src = eexpmap(demo_src, lo_en=lo_en, hi_en=hi_en)
Generating products of type(s) expmap: 100%|██████████| 12/12 [01:21<00:00, 6.78s/it]
Generating products of type(s) expmap: 100%|██████████| 9/9 [00:46<00:00, 5.21s/it]
XGA Ratemaps
Now that we’ve generated images and exposure maps for our weird, custom energy range, we can see that XGA has automatically made RateMaps from those products, I use the individual source as a demonstration here. There are nine ratemaps present because there are three observations of Abell 907, each with three valid instruments:
[7]:
rts = demo_src.get_ratemaps(lo_en=lo_en, hi_en=hi_en)
rts
[7]:
[<xga.products.phot.RateMap at 0x7f8bb5477130>,
<xga.products.phot.RateMap at 0x7f8bae83efa0>,
<xga.products.phot.RateMap at 0x7f8bb5477dc0>,
<xga.products.phot.RateMap at 0x7f8bae7f8430>,
<xga.products.phot.RateMap at 0x7f8bae7f8eb0>,
<xga.products.phot.RateMap at 0x7f8bae83e040>,
<xga.products.phot.RateMap at 0x7f8bae8477c0>,
<xga.products.phot.RateMap at 0x7f8bb54774f0>,
<xga.products.phot.RateMap at 0x7f8bae8384c0>]
Basic properties of ANY XGA Product
Here I just demonstrate some of the most basic (but still useful) properties of XGA’s photometric products. I am going to use one of the ratemaps generated for Abell 907, but the properties I demonstrate in this section will be present in any XGA photometric product. These first properties should actually be present in every XGA product, photometric or otherwise:
[8]:
chosen_ratemap = rts[0]
# If generated using XGA's SAS interface, the product stores the name
# of the source it is associated with
print(chosen_ratemap.src_name, '\n')
# Its also aware of what ObsID and instrument it is from
print(chosen_ratemap.obs_id, chosen_ratemap.instrument, '\n')
# And where the base file is stored
print(chosen_ratemap.path)
A907
0404910601 pn
/home/dt237/code/PycharmProjects/XGA/docs/source/notebooks/tutorials/xga_output/0404910601/0404910601_pn_0.5-10.0keVimg.fits
Basic properties of XGA Photometric products
Here I demonstrate how to access the data and fits header information of an XGA photometric product. I also show off some of the useful functions that are built into photometric objects to make our lives easier.
Accessing an image’s (or in this case a ratemap’s) data is extremely simple, with the added benefit that data is not loaded into memory from the disk until the user actually wants to access it. Its possible to have hundreds of images associated with a single source, so avoiding having data in memory unless its needed helps stop us running out of RAM.
You shouldn’t be concerned that the data shown here is are all zeros, the outskirts of the ratemap are off of the XMM detector and as such have a countrate of zero:
[9]:
# Retrieving the data as a numpy array is as simple as
# calling the data property
my_ratemap_data = chosen_ratemap.data
my_ratemap_data
[9]:
array([[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
...,
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.]])
You can also retrieve the whole header of the fits image by using the header property:
[10]:
chosen_ratemap.header
[10]:
SIMPLE = T / file does conform to FITS standard
BITPIX = 32 / number of bits per data pixel
NAXIS = 2 / number of data axes
NAXIS1 = 512 / length of data axis 1
NAXIS2 = 512 / length of data axis 2
EXTEND = T / FITS dataset may contain extensions
XPROC0 = 'evselect table=/home/dt237/apollo_mnt/xmm_obs/data/0404910601/eclean'
XDAL0 = '0404910601_pn_0.5-10.0keVimg.fits 2021-06-09T16:25:26.000 Create evs'
CREATOR = 'evselect (evselect-3.68) [xmmsas_20190531_1155-18.0.0]' / name of cod
DATE = '2021-06-09T16:25:26.000' / creation date
ORIGIN = 'User site' /
ODSVER = '16.911' / ODS version
ODSCHAIN= 'S2KO' / Processing Chain
FILTER = 'Thin1' / Filter ID
FRMTIME = 199 / [ms] Frame Time, nearest integer
TELESCOP= 'XMM' / Telescope (mission) name
INSTRUME= 'EPN' / Instrument name
REVOLUT = 1184 / XMM revolution number
OBS_MODE= 'POINTING' / Observation mode (pointing or slew)
OBS_ID = '0404910601' / Observation identifier
EXP_ID = '0404910601003' / Exposure identifier
EXPIDSTR= 'S003' / Exposure identifier (Xnnn)
CONTENT = 'EPIC PN IMAGING MODE EVENT LIST' /
DATAMODE= 'IMAGING' / Instrument mode (IMAGING, TIMING, BURST, etc.)
SUBMODE = 'PrimeFullWindowExtended' / Guest Observer mode
OBJECT = 'RXCJ0958-1103' / Name of observed object
DATE-OBS= '2006-05-27T10:58:40' / Start Time (UTC) of exposure
DATE-END= '2006-05-27T15:34:21' / End Time (UTC) of exposure
DATE_OBS= '2006-05-27T09:57:27.000' / Date observations were made
DATE_END= '2006-05-27T15:42:37.000' / Date observations ended
DURATION= 20710.0 / [s] Duration of observation
TIMESYS = 'TT' / XMM time will be TT (Terrestial Time)
MJDREF = 50814.0 / [d] 1998-01-01T00:00:00 (TT) expressed in MJD (
TIMEZERO= 0 / Clock correction (if not zero)
TIMEUNIT= 's' / All times in s unless specified otherwise
CLOCKAPP= T / Clock correction applied
TIMEREF = 'LOCAL' / Reference location of photon arrival times
TASSIGN = 'SATELLITE' / Location of time assignment
OBSERVER= 'Prof Hans Boehringer' / Name of PI
RA_OBJ = 149.592084 / [deg] Ra of the observed object
DEC_OBJ = -11.0597222 / [deg] Dec of the observed object
RA_NOM = 149.592084 / [deg] Ra of boresight
DEC_NOM = -11.0597222 / [deg] Dec of boresight
SRCPOS = 190 / [pixel] Assumed RAWY source position for fast m
SRCPOSX = 36.6997234597762 / [pixel] real-valued RAWX source position
SRCPOSY = 190.216005661132 / [pixel] real-valued RAWY source position
EXPSTART= '2006-05-27T10:58:40' / Start Time (UTC) of exposure
EXPSTOP = '2006-05-27T15:34:21' / End Time (UTC) of exposure
OBSSTART= '2006-05-27T09:57:27.000' / Date observation was started
OBSSTOP = '2006-05-27T15:42:37.000' / Date observation was stopped
ATT_SRC = 'AHF' / Source of attitude data (AHF|RAF|THF)
ORB_RCNS= T / Reconstructed orbit data used?
TFIT_RPD= F / Recalculated signal propagation delays?
TFIT_DEG= 4 / Degree of OBT-MET fit polynomial
TFIT_RMS= 5.43513446215138e-06 / RMS value of OBT-MET polynomial fit
TFIT_PFR= 0.0 / Fraction of disregarded TCS data points
TFIT_IGH= T / Ignored GS handover in TC data?
LONGSTRN= 'OGIP 1.0' /
EQUINOX = 2000.0 / Equinox for sky coordinate x/y axes
RADECSYS= 'FK5' / World coord. system for this file
REFXCTYP= 'RA---TAN' / WCS Coord. type: RA tangent plane projection
REFXCRPX= 25921 / WCS axis reference pixel of projected image
REFXCRVL= 149.591375 / [deg] WCS coord. at X axis ref. pixel
REFXCDLT= -1.38888888888889e-05 / [deg/pix] WCS X increment at ref. pixel
REFXLMIN= 1 / WCS minimum legal QPOE projected image X axis v
REFXLMAX= 51840 / WCS maximum legal QPOE projected image X axis v
REFXDMIN= 1254 / WCS minimum projected image X axis data value
REFXDMAX= 14799 / WCS maximum projected image X axis data value
REFXCUNI= 'deg' / WCS Physical units of X axis
REFYCTYP= 'DEC--TAN' / WCS Coord. type: DEC tangent plane projection
REFYCRPX= 25921 / WCS axis reference pixel of projected image
REFYCRVL= -11.0887222222222 / [deg] WCS coord. at Y axis ref. pixel
REFYCDLT= 1.38888888888889e-05 / [deg/pix] WCS Y increment at ref. pixel
REFYLMIN= 1 / WCS minimum legal QPOE projected image Y axis v
REFYLMAX= 51840 / WCS maximum legal QPOE projected image Y axis v
REFYDMIN= 17644 / WCS minimum projected image Y axis data value
REFYDMAX= 33229 / WCS maximum projected image Y axis data value
REFYCUNI= 'deg' / WCS Physical units of Y axis
AVRG_PNT= 'MEDIAN' / Meaning of PNT values (mean or median)
RA_PNT = 149.591375 / [deg] Actual (mean) pointing RA of the optical
DEC_PNT = -11.0887222222222 / [deg] Actual (mean) pointing Dec of the optica
PA_PNT = 306.720916748047 / [deg] Actual (mean) measured position angle of
HISTORY 1-06-09T16:25:26
HISTORY Created by evselect (evselect-3.68) [xmmsas_20190531_1155-18.0.0] at '
XMMSEP = 83 / 83 primary image keywords follow
SLCTEXPR= '(PI in [500:10000])' / Filtering expression used by evselect
HDUCLASS= 'OGIP' / Format conforms to OGIP/GSFC conventions
HDUCLAS1= 'IMAGE' / File contains an Image
HDUCLAS2= 'TOTAL' / Gross Image
HDUVERS1= '1.1.0' / Version of format
CTYPE1 = 'RA---TAN' / Coord. type: RA tangent plane projection
CRPIX1 = 256.505747126437 / WCS reference pixel
CRVAL1 = 149.591375 / [deg] coord. at X axis ref. pixel
CUNIT1 = 'deg' / Physical units of X axis
CDELT1 = -0.00120833333333333 / WCS pixel size
CTYPE1L = 'X' / WCS coordinate name
CRPIX1L = 1 / WCS reference pixel
CRVAL1L = 3692.0 / WCS reference pixel value
CDELT1L = 87.0 / WCS pixel size
LTV1 = -41.4367816091954 /
LTM1_1 = 0.0114942528735632 /
CTYPE2 = 'DEC--TAN' / Coord. type: DEC tangent plane projection
CRPIX2 = 256.505747126437 / WCS reference pixel
CRVAL2 = -11.0887222222222 / [deg] coord. at Y axis ref. pixel
CUNIT2 = 'deg' / Physical units of Y axis
CDELT2 = 0.00120833333333333 / WCS pixel size
CTYPE2L = 'Y' / WCS coordinate name
CRPIX2L = 1 / WCS reference pixel
CRVAL2L = 3692.0 / WCS reference pixel value
CDELT2L = 87.0 / WCS pixel size
LTV2 = -41.4367816091954 /
LTM2_2 = 0.0114942528735632 /
WCSNAMEL= 'PHYSICAL' / WCS L name
WCSAXESL= 2 / No. of axes for WCS L
LTM1_2 = 0 /
LTM2_1 = 0 /
TIMEDEL = 0.19453132 / [s] Length of exposure entry interval
DSTYP1 = 'CCDNR' / data subspace descriptor: name
DSTYP2 = 'FLAG' / data subspace descriptor: name
DSTYP3 = 'PATTERN' / data subspace descriptor: name
DSTYP4 = 'FLAG' / data subspace descriptor: name
DSTYP5 = 'TIME' / data subspace descriptor: name
DSUNI5 = 's' / data subspace descriptor: units
DSTYP6 = 'PI' / data subspace descriptor: name
DSUNI6 = 'eV' / data subspace descriptor: units
DSVAL1 = '1' / data subspace descriptor: value
DSFORM2 = 'X' / data subspace descriptor: data type
DSVAL2 = 'b00xx00000x0xxxxxxxxxxxxxxxxx' / data subspace descriptor: value
DSVAL3 = ':4' / data subspace descriptor: value
DSVAL4 = '0' / data subspace descriptor: value
DSVAL5 = 'TABLE' / data subspace descriptor: value
DSREF5 = ':GTI00005' / data subspace descriptor: reference
DSVAL6 = '500:10000' / data subspace descriptor: value
2DSVAL1 = '2' / data subspace descriptor: value
2DSREF5 = ':GTI00105' / data subspace descriptor: reference
3DSVAL1 = '3' / data subspace descriptor: value
3DSREF5 = ':GTI00205' / data subspace descriptor: reference
4DSVAL1 = '4' / data subspace descriptor: value
4DSREF5 = ':GTI00305' / data subspace descriptor: reference
5DSVAL1 = '5' / data subspace descriptor: value
5DSREF5 = ':GTI00405' / data subspace descriptor: reference
6DSVAL1 = '6' / data subspace descriptor: value
6DSREF5 = ':GTI00505' / data subspace descriptor: reference
7DSVAL1 = '7' / data subspace descriptor: value
7DSREF5 = ':GTI00605' / data subspace descriptor: reference
8DSVAL1 = '8' / data subspace descriptor: value
8DSREF5 = ':GTI00705' / data subspace descriptor: reference
9DSVAL1 = '9' / data subspace descriptor: value
9DSREF5 = ':GTI00805' / data subspace descriptor: reference
10DSVAL1= '10' / data subspace descriptor: value
10DSREF5= ':GTI00905' / data subspace descriptor: reference
11DSVAL1= '11' / data subspace descriptor: value
11DSREF5= ':GTI01005' / data subspace descriptor: reference
12DSVAL1= '12' / data subspace descriptor: value
12DSREF5= ':GTI01105' / data subspace descriptor: reference
ONTIME01= 6050.0 / Sum of GTIs for a particular CCD
ONTIME02= 6050.0 / Sum of GTIs for a particular CCD
ONTIME03= 6050.0 / Sum of GTIs for a particular CCD
ONTIME04= 6050.0 / Sum of GTIs for a particular CCD
ONTIME05= 6050.0 / Sum of GTIs for a particular CCD
ONTIME06= 6050.0 / Sum of GTIs for a particular CCD
ONTIME07= 6050.0 / Sum of GTIs for a particular CCD
ONTIME08= 6050.0 / Sum of GTIs for a particular CCD
ONTIME09= 6050.0 / Sum of GTIs for a particular CCD
ONTIME10= 6050.0 / Sum of GTIs for a particular CCD
ONTIME11= 6050.0 / Sum of GTIs for a particular CCD
ONTIME12= 6050.0 / Sum of GTIs for a particular CCD
EXPOSURE= 6050.0 / max of ONTIMEnn values
And a specific entry by indexing the header object using the entries name:
[11]:
chosen_ratemap.header['OBSERVER']
[11]:
'Prof Hans Boehringer'
Converting coordinates with XGA Photometric products
Reading in the header means that XGA has access to the World Coordinate System (WCS) information of the fits image, and using this I wrote a function that will convert between different coordinate systems for a given photometric product. Again, any coordinates must be passed in as Astropy quantity objects:
[12]:
# Defining a position to convert in pixel coordinates
pix_coord = Quantity([100, 100], 'pix')
# The pixel coordinate can be converted to degrees like this
print(chosen_ratemap.coord_conv(pix_coord, 'deg'), '\n')
# If the product fits file had a WCS entry for the XMM sky coordinates system,
# then we can convert to that as well
print(chosen_ratemap.coord_conv(pix_coord, 'xmm_sky'))
[149.782975 -11.27656289] deg
[12392. 12392.] xmm_sky
We can also actually define an Astropy quantity in XMM’s sky units, by dint of a custom unit defined in the XGA utils file (I imported it at the top of this tutorial). Please be aware that the Astropy module is not aware of this custom unit, so defining a quantity with a string representation of the unit will not work:
[13]:
# Defining an XMM Sky position
sky_coord = Quantity([15000, 15000], xmm_sky)
# Converting to pixels
print(chosen_ratemap.coord_conv(sky_coord, 'pix'), '\n')
# Converting to an RA-Dec position
print(chosen_ratemap.coord_conv(sky_coord, 'deg'))
[130 130] pix
[149.74602109 -11.24036252] deg
We can also easily convert between any combination of these units, and here I demonstrate how to convert the user supplied coordinates (used when the cluster was defined), as well as the peak coordinates found during declaration:
[14]:
# This is an easy way to access the coordinates the user passed in when declaring the source
print('The initial coordinates were', demo_src.ra_dec)
print(chosen_ratemap.coord_conv(demo_src.ra_dec, 'pix'), '\n')
# And an equally easy way to access the peak coordinates
print('The peak coordinates are', demo_src.peak)
print(chosen_ratemap.coord_conv(demo_src.peak, 'pix'), '\n')
The initial coordinates were [149.59209 -11.05972] deg
[255 280] pix
The peak coordinates are [149.59251341 -11.06395832] deg
[255 276] pix
Viewing XGA Photometric products
One of the most useful features built into XGA’s photometric products is the ability to easily visualise them, with multiple ways to modify the image produced. This includes changing the size of the image, altering the scaling applied (log-scaling is used by default), applying masks, zooming in specific parts of the image, and adding crosshairs to indicate a coordinate.
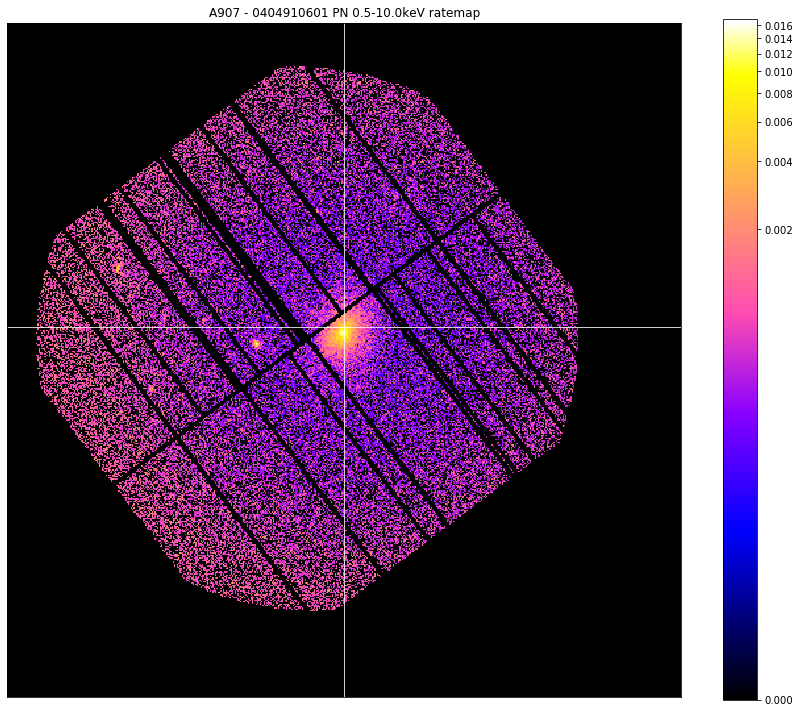
Again, we’re using a ratemap taken from Abell 907, and with one simple line of code we can produce a useful visualisation. I’ve increased the size of the image so that it looks better in the documentation, and used the crosshairs to highlight the coordinate of the user supplied position. Only one crosshair can be placed on an image, but the coordinate can be in degrees, pixels, or XMM sky coordinates:
[15]:
# Displaying the ratemap, with a crosshair for a particular coordinate
chosen_ratemap.view(cross_hair=demo_src.ra_dec, figsize=(12, 10))
Retrieving a value at a certain coordinate
At some point you may wish to know the value of a photometric product at a certain coordinate, a ratemap or an exposure map for instance. There are built in methods to help you retrieve those value which will accept coordinates in any valid unit (degrees, pixels, or XMM sky). The value is returned as an Astropy quantity, with the correct units for the type of product.
I’ll also show you a useful feature of ratemap, that it stores links to the image and expmap that were used to create it, so accessing those constituent products is as easy as using the ‘image’ or ‘expmap’ properties of the ratemap:
[16]:
# For a ratemap, we use the method 'get_rate'
print(chosen_ratemap.get_rate(demo_src.ra_dec), '\n')
# If we wished to know the exposure time at that position, we could access the ratemap's
# expmap and use 'get_exp'
print(chosen_ratemap.expmap.get_exp(demo_src.ra_dec), '\n')
0.006222707412880577 ct s
4981.75439453125 s
Combined images and exposure maps
Now that we have generated images and exposure maps for all the ObsIDs and instruments associated with all these sources that we’re interested in analysing, we will want to bring all that information together by creating combined images and exposure maps. Most XGA analyses take place on combined products, because that ability to find all relevant data for a particular source is one of XGA’s key strengths. Currently I use the SAS routine emosaic to generate the combined data products, and it is nearly as simple as generating images and exposure maps, the main difference is that you need to tell emosaic what type of product you want to combine:
[17]:
# ------------IMAGES------------
# Starting off with making combined images for the sample
demo_smp = emosaic(demo_smp, 'image', lo_en=lo_en, hi_en=hi_en)
# And for the single source
demo_src = emosaic(demo_src, 'image', lo_en=lo_en, hi_en=hi_en)
# -----------EXPMAPS------------
# The same again but telling emosaic that we want combined exposure maps this time
demo_smp = emosaic(demo_smp, 'expmap', lo_en=lo_en, hi_en=hi_en)
demo_src = emosaic(demo_src, 'expmap', lo_en=lo_en, hi_en=hi_en)
Generating products of type(s) image: 100%|██████████| 4/4 [00:00<00:00, 9.67it/s]
Generating products of type(s) image: 100%|██████████| 1/1 [00:00<00:00, 2.51it/s]
Generating products of type(s) expmap: 100%|██████████| 4/4 [00:00<00:00, 10.10it/s]
Generating products of type(s) expmap: 100%|██████████| 1/1 [00:00<00:00, 2.52it/s]
I’m also going to read out the combined ratemap into a variable for easy use in other parts of this tutorial:
[18]:
chosen_comb_rt = demo_src.get_products('combined_ratemap', extra_key='bound_0.5-10.0')[0]
Retrieving and using masks from a source
It is common practise in photometric analyses to mask out any sources that might bias or interfere with the source that you are investigating, and as such each XGA source object is able to produce masking arrays. These masking arrays are filled with ones and zeros, where any zero elements are to be considered ‘masked’, and masked image/expmap/ratemap data can be calculated by multiplying the image/expmap/ratemap data by the masking array.
XGA is capable of producing several types of mask; interloper masks (where non-relevant sources are masked), source masks (where any part of the product not within a certain source region is masked out), and standard masks (which are a combination of the last two types).
An XGA source can generate masks for any of the ObsIDs associated with it, which is important because the ObsIDs are all likely to have different pointing coordinates and roll angles, and as such the pixel coordinates will not correspond to the same RA-Dec coordinates. If no ObsID information is supplied then XGA will assume that the mask should be generated for a combined photometric product.
I’m also going to demonstrate the masking functionality of the view() method for photometric XGA products by showing the effects of the masks I’m generating on actual ratemaps:
[19]:
# This generates a mask that removes interloper sources (any non-relevant source). As we
# didn't provide an ObsID it is only valid for the combined products
interloper_mask = demo_src.get_interloper_mask()
# It is very easy to apply the mask to remove any interlopers from the ratemap
chosen_comb_rt.view(cross_hair=demo_src.ra_dec, mask=interloper_mask, figsize=(12, 10))
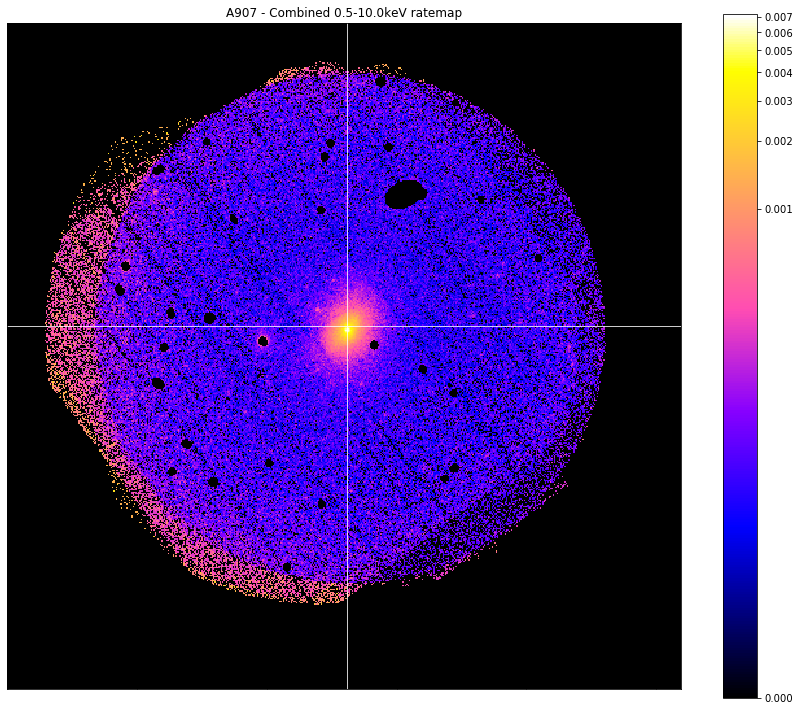
Its just as easy to produce an interloper mask for a specific ObsID, and though the ratemaps look extremely similar, please note that this is because the three observations associated with this source all have near identical pointing coordinates. That means that the shape of this image is very similar to the 512x512 of the individual images. This may not be the case for other sources, where separate observations added together can result in quite different image shapes.
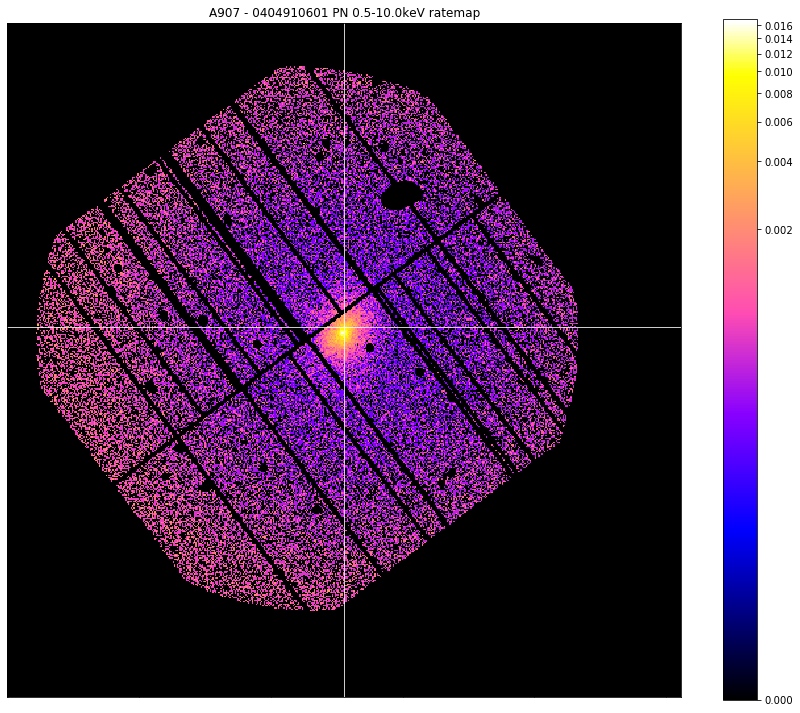
Here we produce a mask for the Abell 907 ratemap we retrieved earlier:
[20]:
# Much the same process as to generate the mask for the combined ratemap, but we also supply an ObsID
individual_interloper_mask = demo_src.get_interloper_mask(obs_id=chosen_ratemap.obs_id)
# It is very easy to apply the mask to remove any interlopers from the ratemap
chosen_ratemap.view(cross_hair=demo_src.ra_dec, mask=individual_interloper_mask, figsize=(12, 10))
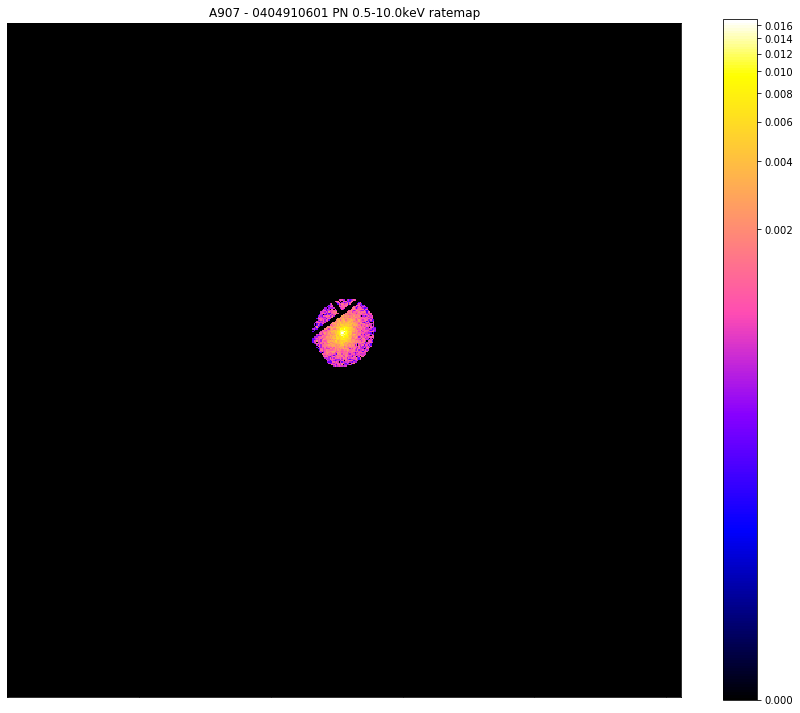
Now we are going to learn how to generate masks that lets us select the source region of interest. At this point we have to make a choice as to which source region we’re interested in; most source objects will allow us to generate a mask from the region files which were supplied to XGA in the configuration file, but if you have a point source then you could also choose ‘point’, and if you have a Galaxy Cluster then you could also select one of the overdensity regions (such as ‘r200’).
When you generate a source region mask, you will also be supplied with a matching background region mask, which is a region extending between back_inn_rad_factor - back_out_rad_factor multiplied by the radius of interest, where these keyword arguments are set when defining a source object.
[21]:
# A source and background mask generated for the selected source from our region file for
# this particular ObsID
region_masks = demo_src.get_source_mask('region', chosen_ratemap.obs_id)
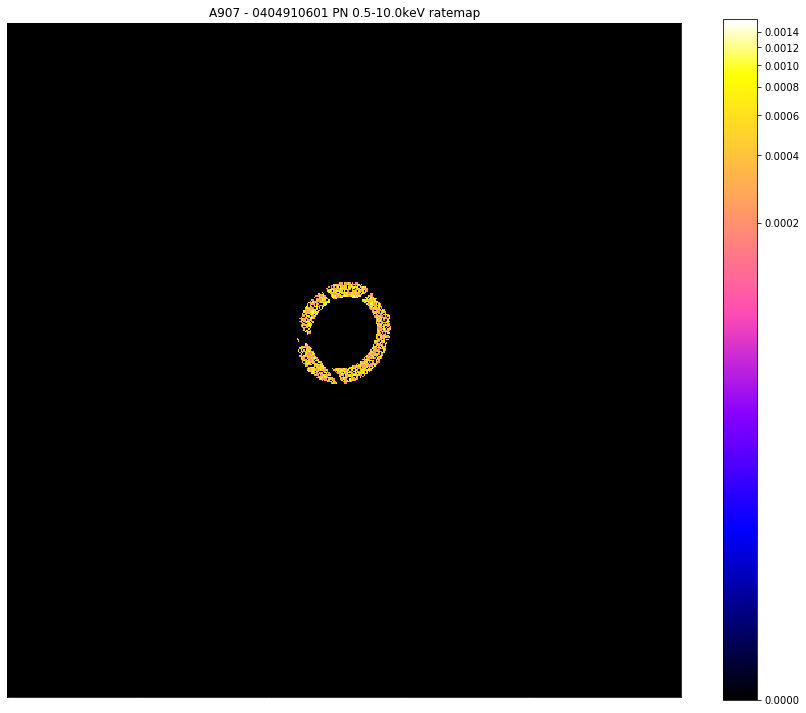
# Here I will show the source mask applied to the 0404910601-PN ratemap in the 0.5-10.0keV range
chosen_ratemap.view(mask=region_masks[0], figsize=(12, 10))
# I will also show the background mask applied to this ratemap
chosen_ratemap.view(mask=region_masks[1], figsize=(12, 10))
/home/dt237/code/PycharmProjects/XGA/xga/sources/base.py:1334: UserWarning: You cannot use custom central coordinates with a region from supplied region files
warnings.warn("You cannot use custom central coordinates with a region from supplied region files")
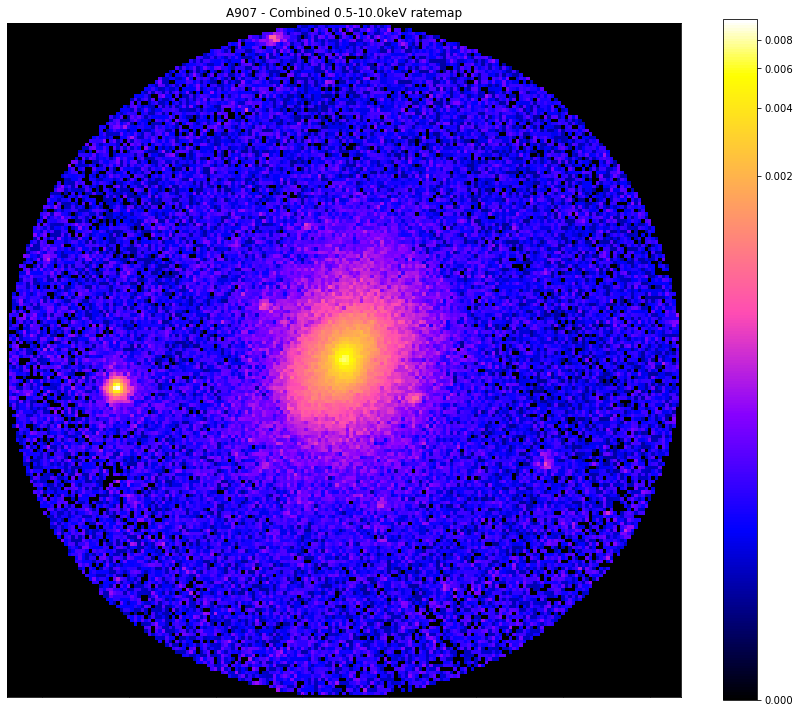
Now we’ve seen how to access a region file region, we’re going to look at an R\(_{500}\) region, this also seems like a good time to demonstrate XGA’s automatic zoom functionality when viewing photometric products:
[22]:
# A source and background mask generated for the combined ratemap for Abell 907 at R_500
r500_masks = demo_src.get_source_mask('r500')
# Here I will show the source mask applied to the combined ratemap in the 0.5-10.0keV range
chosen_comb_rt.view(mask=r500_masks[0], figsize=(12, 10), zoom_in=True)
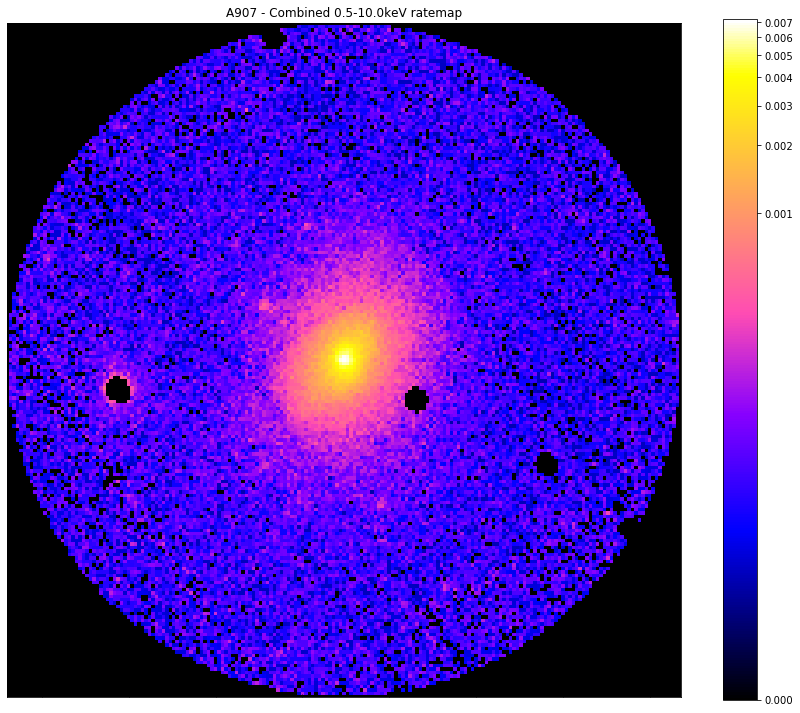
Finally, we can still see that there are some obvious point sources remaining in this source region, and for a real analysis we would want to be able to remove them. As such we use the source method ‘get_mask’, and apply it to the same combined ratemap.
I also wish to note that the user does not have to generate these masks centered at the default central coordinates for the source object in question, you can also specify another central coordinate using central_coord in a mask generation method.
When we see the image below we can quite clearly see examples of a few point sources that obviously have to be removed before a meaningful analysis of the galaxy cluster can begin:
[23]:
# A source and background mask generated for the combined ratemap for Abell 907 at R_500
r500_interloper_masks = demo_src.get_mask('r500',)
# Here I will show the source mask applied to the combined ratemap in the 0.5-10.0keV range
chosen_comb_rt.view(mask=r500_interloper_masks[0], figsize=(12, 10), zoom_in=True)
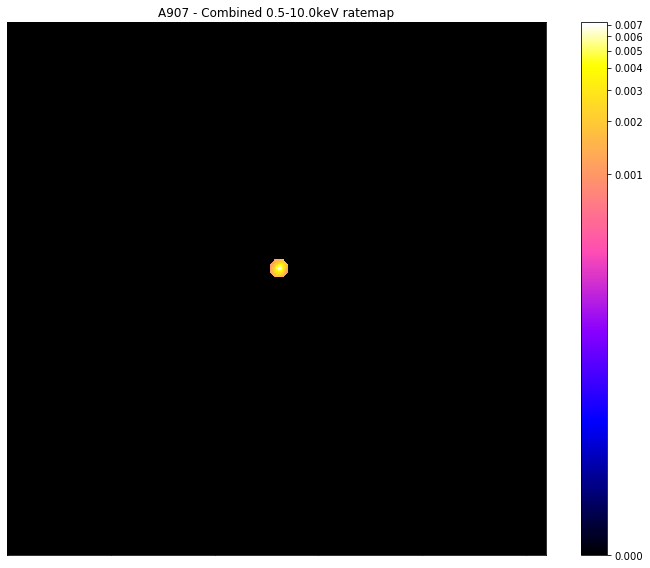
Creating a custom mask
You can also create a mask with whatever custom radius you desire, suitable to be applied to either the merged images or one of the individual observation images. You may also define custom coordinates to centre the new mask on, as well as defining both inner and outer radii (if you wish to make an annulus). I shall start by making a mask with an outer radius of 0.01 degrees (though if the source in question has redshift information you can also use a proper distance unit) for the combined image of the demo source:
[24]:
custom_mask = demo_src.get_custom_mask(Quantity(0.01, 'deg'))
custom_mask
[24]:
array([[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
...,
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.]])
[25]:
chosen_comb_rt.view(mask=custom_mask)
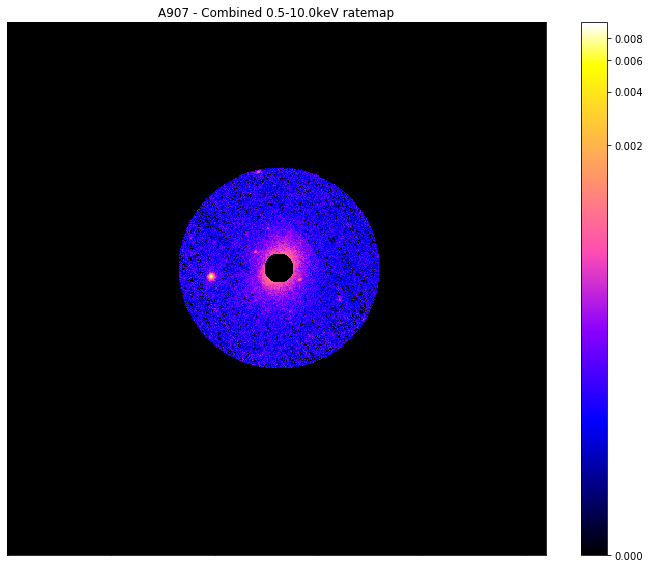
Now I shall make a mask with the core of this galaxy cluster excised (the mask shall allow everything from 0.15-1R\(_{500}\)), but also I shall decide that I no longer want to remove interloper sources:
[26]:
custom_mask = demo_src.get_custom_mask(demo_src.r500, demo_src.r500*0.15, remove_interlopers=False)
chosen_comb_rt.view(mask=custom_mask)
Measuring signal-to-noise
Measuring the signal to noise of a given source is a fairly common task in astronomy, and an ability which has been implemented and added to the photometric product classes in XGA. It is a simple matter to quickly find the signal to noise of a galaxy cluster within an overdensity radius for instance, you need only call the info() method. There you will find that XGA has automatically measured the signal-to-noise within the supplied overdensity radii:
[27]:
demo_src.info()
-----------------------------------------------------
Source Name - A907
User Coordinates - (149.59209, -11.05972) degrees
X-ray Peak - (149.59251340970866, -11.063958320861634) degrees
nH - 0.0534 1e+22 / cm2
Redshift - 0.16
XMM ObsIDs - 3
PN Observations - 3
MOS1 Observations - 3
MOS2 Observations - 3
On-Axis - 3
With regions - 3
Total regions - 69
Obs with one match - 3
Obs with >1 matches - 0
Images associated - 27
Exposure maps associated - 27
Combined Ratemaps associated - 2
Spectra associated - 0
R200 - 1700.0 kpc
R200 SNR - 230.23
R500 - 1200.0 kpc
R500 SNR - 251.61
-----------------------------------------------------
If, however, you wish to retrieve the signal to noise programmatically (or measure it within a custom radius), you can simply use the get_snr method, which I hear use to retrieve the signal to noise of this cluster within the R\(_{500}\) and within a custom radius of 300kpc:
[28]:
r500_snr = demo_src.get_snr('r500')
print(r500_snr)
cust_snr = demo_src.get_snr(Quantity(300, 'kpc'))
print(cust_snr)
251.61136957214998
206.61225573609303
You are also able to perform a simple assessment of which observation associated with a given source is ‘the best’, by ranking them by signal to noise of the source. Call the snr_ranking method, supply it with a radius within which to measure the SNR, and it will return an array of ObsID-instrument combinations, and an array of SNR values, going from worst to best:
[29]:
oi_combs, snr_vals = demo_src.snr_ranking('r500')
[30]:
oi_combs
[30]:
array([['0201901401', 'mos1'],
['0201901401', 'mos2'],
['0201901401', 'pn'],
['0201903501', 'mos1'],
['0201903501', 'mos2'],
['0404910601', 'mos2'],
['0404910601', 'mos1'],
['0404910601', 'pn'],
['0201903501', 'pn']], dtype='<U10')
[31]:
snr_vals
[31]:
array([ 71.88691358, 71.90096549, 88.79940544, 94.18597822,
96.22761135, 108.10871705, 108.73900404, 114.58252691,
124.73613614])
Bulk generation of photometric products
If you wished to generate images and exposure maps in a particular energy range for a large number of unrelated observations, then you would use a NullSource object.
Normally, when defining a NullSource object, it would automatically associate every available ObsID with itself, but it is possible to pass a list of specific ObsIDs of interest (which I do in this demo so I don’t destroy my laptop):
[32]:
# Four randomly select observation that will make up the NullSource
chosen_obs = ['0677630133', '0301900401', '0770581201', '0827201501']
# And defining the source with four unrelated observations associated with it
bulk_src = NullSource(chosen_obs)
[33]:
# If we run the standard info() method then we can say a very little information about the source
bulk_src.info()
-----------------------------------------------------
Source Name - 4Observations
XMM ObsIDs - 4
PN Observations - 4
MOS1 Observations - 4
MOS2 Observations - 4
-----------------------------------------------------
Now we can generate images and exposure maps for this bulk sample, though we cannot create combined data products through the emosaic tool because it makes no sense - these are four random observations and have no common coordinate on which they could all be combined:
[34]:
# And of course we can generate images and exposure maps in the same way as with the other sources
evselect_image(bulk_src, lo_en=lo_en, hi_en=hi_en)
eexpmap(bulk_src, lo_en=lo_en, hi_en=hi_en)
Generating products of type(s) image: 100%|██████████| 12/12 [00:07<00:00, 1.60it/s]
Generating products of type(s) ccf: 100%|██████████| 4/4 [00:12<00:00, 3.08s/it]
Generating products of type(s) expmap: 100%|██████████| 12/12 [00:59<00:00, 4.96s/it]
[34]:
<xga.sources.base.NullSource at 0x7f8bae415880>